- The chickpea gene expression atlas could help deploy genetic information to raise productivity and boost climate resilience in chickpea and other food legumes.
- The atlas can be applied for identification of genes that are potentially involved in conferring tolerance/resistance to production constraints such as drought.
- Chickpea together with other crops such as finger millet, groundnut are often branded as forgotten “orphan crops” as they tend to receive less attention from science although they are crucial in providing income to the poorest farmers and are staples in local diets. There is a global demand for chickpeas as consumers become aware of sustainable consumption.
The humble chickpea, often sidelined as an “orphan” crop, is not short of attention anymore, thanks to the “omics” era and a growing awareness on intake of sustainable and non-processed foods.
Stomach this: a rising appetite for hummus, the iconic mashed chickpea dish, has pushed countries to plant record acreage of the crop. Meanwhile a drought-linked shortage in chickpea harvest in India — world’s largest chickpea producing country — led to a spike in prices of the popular Middle Eastern dish in the United Kingdom.
Now, the chickpea (Cicer arietinum) has got its own genetic atlas following a five-year long effort by researchers from International Crops Research Institute for the Semi‐Arid Tropics (ICRISAT), Hyderabad, India.
The researchers say that genetic information mapped in the atlas could be harnessed to boost productivity and climate resilience in chickpea and other food legumes.
The Cicer arietinum gene expression atlas (CaGEA) is a comprehensive catalogue that provides data on when (timing) and where (site) a particular gene on the genome will be expressed during chickpea plant growth.
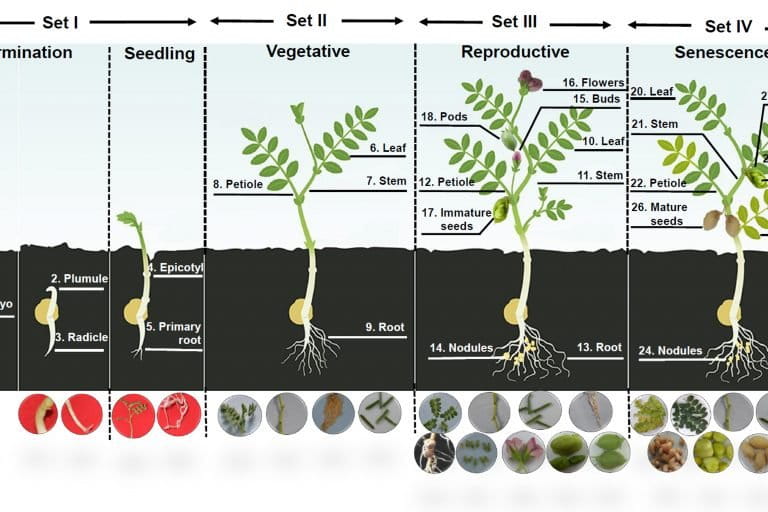
The atlas charts out genetic bottlenecks that limit crop yield during different plant developmental stages.
Plant growth and development are controlled by programmed expression of a suite of genes at the given time, stage and tissue. Gene expression is the generation of a functional gene product from the information encoded by a gene, through the processes of transcription and translation.
Information on gene expression patterns is crucial in understanding how the underlying genome sequence translates into specific observable characteristics at key developmental stages.

“The gene expression atlas could predict cluster of genes expressed in each tissue at different plant developmental stages. Hence, we developed genome-wide gene expression atlas in chickpea to understand key genetic bottlenecks that limit crop yield under different plant developmental process,” Himabindu Kudapa, the first author of the study, told Mongabay-India.
“Genetic bottleneck stems from a dearth of genetic/natural variation for a given trait (characteristic),” said Rajeev K. Varshney, who is the director of the Genetic Gains Research Programme at ICRISAT and also the director for the Gene Expression Atlas project.
“For instance, when a particular crop is exposed to one particular disease, we need to have some line/variety that has resistance to this particular disease. If you don’t have this kind of variation, we may not be able to develop better varieties to address the issue,” Varshney explained.
What’s so special about chickpeas?
Chickpea is the world’s second most widely grown legume. With its capacity for biological nitrogen fixation, it replenishes soil fertility, diminishes the need for added nitrogen fertilisers and is a source of high quality dietary vegetarian protein.
Originating in southeast Turkey and Syria, chickpea is known as one of the founder crops of modern agriculture. It is grown mostly in south Asia and sub-Saharan Africa, which accounts for more than 75 percent of the world’s chickpea area.
There are two main types of chickpeas: small-seeded “desi” and larger-seeded “kabuli”. Consumption of desi is restricted primarily to some parts of Asia, whereas kabuli is a popular and valuable global commodity, the researchers said.
Due to domestication and breeding, chickpea has a narrow genetic base and so it doesn’t have a lot of resistance and tolerance to diseases, they observed.

It is mostly cultivated in semi-arid environments and on soils of poor agricultural quality.
This combined with its “susceptibility to drought and debilitating fungal diseases” has “restricted yields to less than one tonne/hectare, which is considerably below the theoretical potential.”
According to Indian Council of Agricultural Research, despite contributing more than 70 percent to the global chickpea area and production, India remains a net importer of chickpea due to high national demand. Although significant yield increases have been reported under experimental research conditions, farm-level increases have been more modest.
“The atlas will be very useful for identification of genes that are potentially involved in imparting tolerance/resistance to production constraints such as drought. Subsequently, these genes can be used in genomics-assisted breeding for developing better varieties with higher productivity,” Kudapa suggested.
In the present study, the chickpea gene expression atlas has been created from a widely used drought tolerant cultivar ICC 4958.
“Now we have better knowledge about the genes associated with yield under drought stress. This will help in developing climate-resilient chickpea varieties to provide better produce to small holder famers. Our work will help India towards achieving its goal of self-sufficiency in pulses,” observed Varshney.

In order to ensure self-sufficiency, the pulse requirement in the
country is projected at 39 million tonnes by the year 2050 which
necessitates an annual growth rate of 2.2 percent, states the ‘Vision 2050′ document crafted by ICAR-Indian Institute of Pulses Research.
The Vision 2050 calls for augmenting pulses production in the country through technology upgradation and targets major research and development work in areas such as low genetic yield potential, poor, unstable yield, huge post-harvest losses, inadequate adoption of improved technologies and low profitability.
The plan talks about tackling these challenges through integration of conventional approaches with cutting edge research in areas including genomics, molecular marker assisted breeding, transgenics, molecular approaches for stress management and resource conservation.
Varshney added that the atlas was developed using the RNA-sequence approach by generating gene expression data from 27 different tissues representing five developmental stages covering entire life cycle of the chickpea.
“The deployment of the knowledge from the gene expression atlas together with the developed genomic resources including chickpea genome sequence in crop improvement programs would enhance the precision and efficiency in chickpea breeding programs,” Varshney said.
Orphan no more
The story behind the atlas’s genesis began with decoding of the genome sequence of chickpea in 2013.
“While genome sequencing of chickpea provided the genes to us, we didn’t know the stage and tissue associated with expression of genes, as all the genes are not expressed in all tissues and all at the time. Therefore, we started developing gene expression atlas of chickpea,” Varshney recalled.
Chickpea together with other crops such as finger millet, groundnut are often branded as “orphan crops” as they tend to receive less attention from science although they are crucial in providing income to the poorest farmers and are staples in local diets.
“Chickpea used to be called an orphan crop, as there are not many researchers around the world working on this crop. One of the reasons is this crop is important in India and several African countries as it is used for food and nutrition security, but it is not that important in the western world,” Varshney said.
As a result, chickpea crop remained untouched from the genomic revolution until 2005, the scientist lamented.
“With an objective to use the genomics-assisted breeding, we started to develop genomic resources,” the researchers said.
Brent Kaiser, professor of Legume Biology at the University of Sydney , who was not associated with the study, said the atlas has the scope to expand with more sequencing.
“Having a gene atlas is really useful. Having expression profiles across tissues is useful for recording which genes are expressed, where and when. This paper and the data represent the first iteration of that atlas. And as they sequence more and have more expression pattern data, the atlas will grow,” Kaiser told Mongabay-India.
Kaiser is leading projects that focus on developing heat-tolerant chickpea lines and the identification and development of rhizobial inoculants for northern Australian soils.
“We grow a lot of chickpea here. It is a legume of choice for a lot of growers here. Australia has a very large grains industry so legumes are used in rotation. Over the last 10 years chickpea has become more and more favourable as a rotation crop. Genetics are getting better,” Kaiser said.
He noted that by 2020, population growth in India is expected to surpass China, and given the nation’s appetite for legumes, ramping up chickpea production offers huge potential for Australian growers.
“We export about 85 to 90 percent of what we grow and most of that goes to India. India grows eight times more chickpea than we do. There is a growing demand for this crop and India is leading that demand. India needs another one-and-a-half million tonnes of pulses (mainly chickpeas) per year for the next 15 years to meet their expected population growth food requirements,” Kaiser explained.
Apart from the business opportunities, Kaiser said if farmers are encouraged to plant chickpeas or other pulses then we can significantly reduce our ecological footprint.
“People are aware socially of what they are eating now. They want high quality, sustainable, green eco-friendly types of crops and there is a lot of opportunity,” Kaiser signed off.
CITATION:
Kudapa, H., Garg, V., Chitikineni, A., & Varshney, R. K. (2018). The RNA‐Seq‐based high resolution gene expression atlas of chickpea (Cicer arietinum L.) reveals dynamic spatio‐temporal changes associated with growth and development. Plant, cell & environment.
BANNER IMAGE: Green chickpeas. Photo by Jorge Royan/Wikimedia Commons.













